On Friday, I headed uptown to attend the Miami Breast Cancer Conference (#MBCC14) held at the Fontainebleau Hotel and organised by the Physicians Education Resource (PER). It was fun to grab a local Deco Bike and furiously cycle over 45 blocks in under half an hour – most probably the only attendee who arrived on two wheels that day!

MBCC14: Dr Lance Liotta
Now, I haven’t attended this event since it was at the Loews Hotel in midtown, which was rather low key and fairly small. Certainly there wasn’t a big exhibition area then, as far I can recall. Fast forward a decade on and the event is MUCH bigger, with an excellent Academic panel and an interesting mix of didactic talks and case studies. The stage setting is also much more impressive, as you can see in the photo right.
To give you some basic background, the audience polls at the beginning of the first day were really useful to put things into context:
- The majority of attendees (88%) were physicians (mix of Community medical oncologists, radiation oncologists and surgical oncologists)
- 49% of respondents treated 1–5 patients with breast cancer per week
- 25% of respondents treated 6–10 patients with breast cancer per week
Being a scientist, and having missed the San Antonio Breast Cancer Symposium (SABCS) due to an overlap with the American Society of Hematology (ASH) meeting in December, I was particularly keen to catch up on the new developments in genomics and molecular profiling, with early morning talks from Drs Lance Liotta (George Mason Univ) and Debu Tripathy (USC). There were also updates on neoadjuvant treatment for breast cancer by Drs Kathy Albain (Loyola) and Hal Burstein (Dana Farber). Neoadjuvant therapy prior to surgery is an area that is seeing many new trials and potential therapies emerge.
In today’s post, the attention is on the important topic molecular profiling. This is something I believe we will see much more of going forward. Two separate articles will follow on personalised treatment in advanced breast cancer (including TNBC) and also on neoadjuvant developments.
Genomics can sometimes be a bit of a dry topic, at least to some people, as anyone who has sat through slide after slide of those fuzzy green-red assays in systems biology sessions at AACR will attest. This time, much to my pleasant surprise, it was different…
What I heard blew my mind and changed the way I think about some aspects of breast cancer.
Now I’m not joking or trying to hype progress here, but sometimes you experience an epiphany when you least expect it.
To read more about this revelation, you can sign up or sign in below.
This content is restricted to subscribers
Lance Liotta always gives well organised presentations and illustrates the key facts on proteomics (a tough subject for many to follow) with critical learning points. At this year’s MBCC, he focused his talk on a vision for combining genomic mapping with proteomic analysis of the metastatic lesion. Part of the challenge with using genomics is the sheer heterogeneity and complexity of every single patient’s tumour. He also had a second talk in the afternoon that was equally interesting, but more about that in another post.
One of the main findings from the SideOut trial (run by TGen and George Mason, sponsored by the Side Out Foundation; reported at ASCO 2013 – download the poster here) a proof-of-concept study, which showed that molecular profiling often yielded a treatment recommendation that was different from the one recommended by the treating physician:
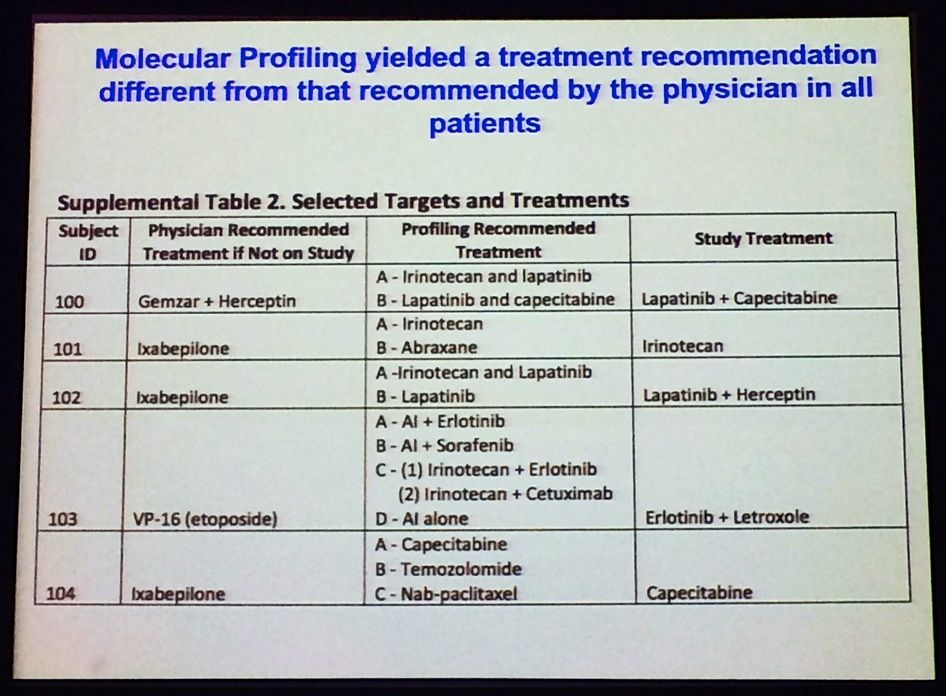
Source: L Liotta, MBCC
You can see that some of the regimens mentioned here are quite noticeably different – patient #103 is particularly fascinating, for example.
Here’s my quick summary of some of the main points from his first talk:
- Basic concept of SideOut I: map the signaling network of metastatic tumour cells to understand which growth or survival pathways are functionally in use in the tissue microenvironment.
- Combine this information with genomic analysis from biopsies to determine true drivers from passengers.
- Use a combination of genomics and proteomics to recommend appropriate therapies.
- The trial was largely successful at generating good responses to therapy and demonstrating PFS – 40% of patients exceeded the PFS ratio of 1.3 and three pats still continue on therapy for 199, 254 and 816 days.
- 60% of patient samples had activation of drug targets in only 3 major clusters i.e.
- pan-HER-AKT
- EGFR/Src/ERK/mTOR
- IGF/RAF/MEK/PLK1
- Improved treatment may therefore be facilitated by biomarker-led understanding of subgroup molecular targets, which may predict benefit from currently approved agents and newer targeted drugs.
- Subclones are selected out based on selective pressure i.e. survival in a secondary tissue or organ during metastasis or survival in the face of therapy (adaptive resistance). This is something we need to learn more about as our knowledge of the biology of the disease improves.
Following the success of this trial, SIDEOUT II has now opened in 9 sites looking at metastatic breast cancer patients progressing after 1–3 lines of therapy. The study will investigate genome sequencing, protein pathway mapping and multiplexed IHC before using all of the information available to provide a molecular rationale for individualising therapy.
For the SideOut I study, Liotta gave a nice example of a typical patient case study, as shown below. The idea was to illustrate how they investigators tackled this difficult case and used the genomic and proteomic data to make better clinical decisions. Note the patient had TNBC, yet had different findings for HER2 status based on two different tests – this isn’t an uncommon finding with lab results, unfortunately:
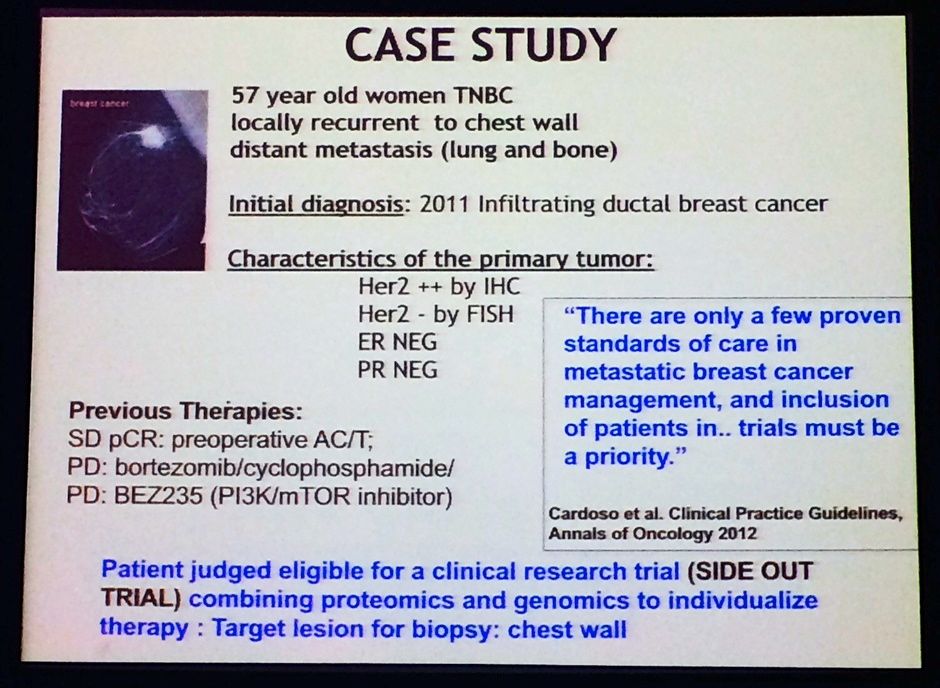
Source: L Liotta, MBCC
After the proteomic and genomic analysis, this is what they ended up with. Note the recommended treatment regimen that resulted – not something you would normally consider with such a detailed work-up!
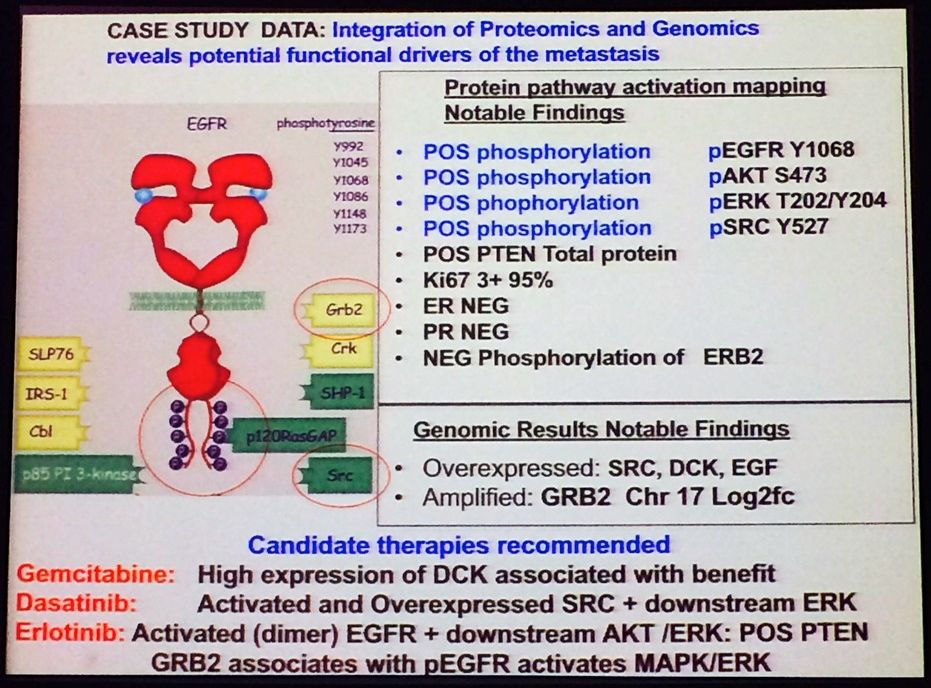
Source: L Liotta MBCC
To put this in better context – consider the attendee poll on what tests the physicians would order based on the biopsy of a metastatic lesion suggested that 60% would run ER, PR and HER2 only, while less than a third would test for ER, PR, HER2 and genomic profiling. Personally, I was really surprised that so few respondents would consider genomic sequencing in the metastatic setting given the sheer molecular complexity that exists. Clearly, there is a molecular world beyond hitting ER, PR and HER2.
Ultimately, the proof of the pudding in any clinical trial is outcome – how well did the patients do when molecular profiling was used to guide therapy? Remember that many of these patients had quite advanced disease and were considered difficult to treat.
The answer is quite well, as this waterfall plot demonstrates:
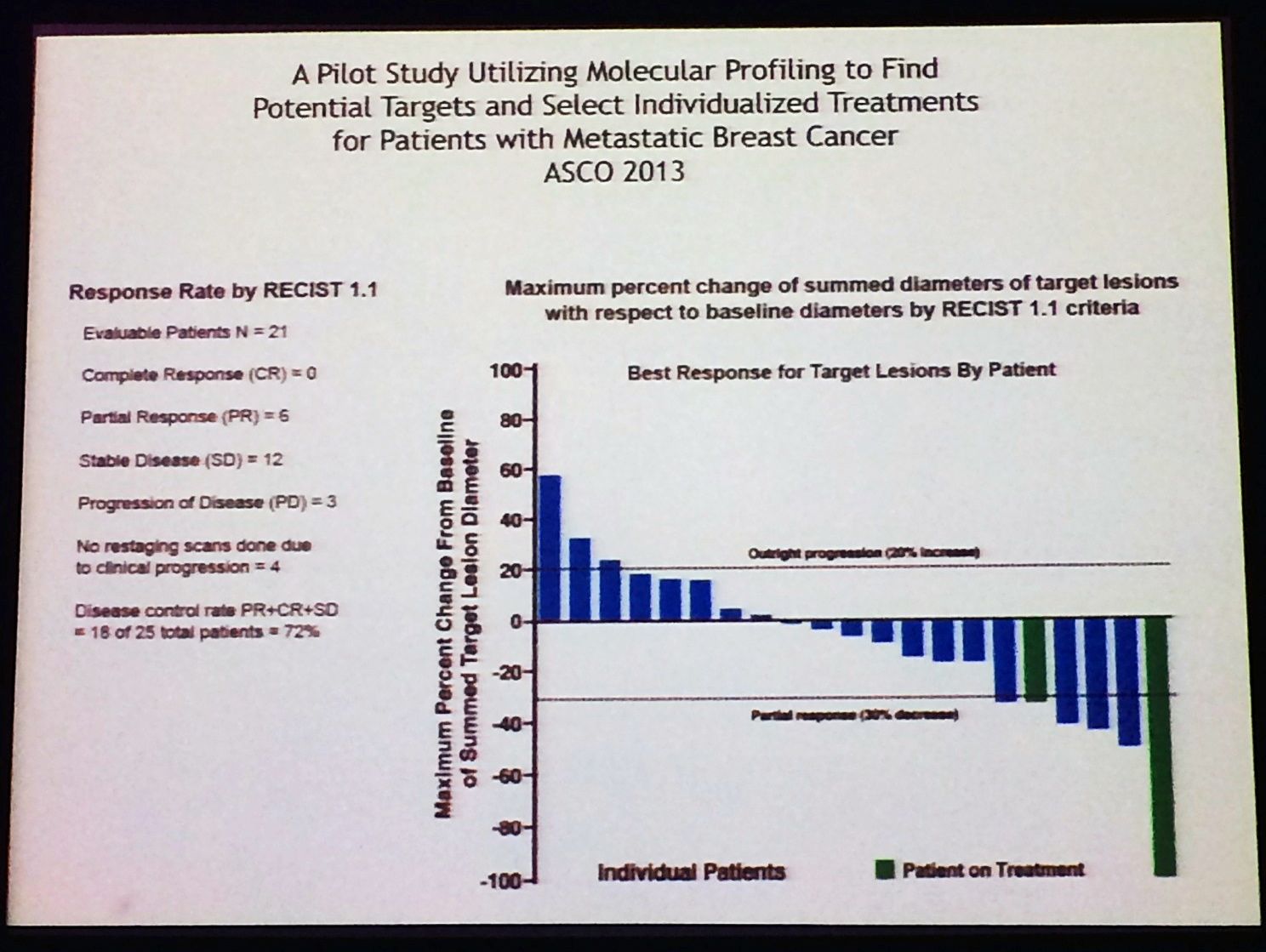
We have to wonder how can we possibly expect to treat any patient successfully, if we don’t know what driver mutations and targets exist? In this respect, lung cancer has truly come out of the shadows and leap-frogged breast cancer in terms of molecular profiling and targeted therapies, at least in Academia.
In the future, it may be possible to better define triple negative breast cancers (TNBC) by what the are, rather than what they’re not i.e. ER, PR, HER2 negative, which is a broad catch-all and a very heterogenous population indeed.
Meanwhile, tomorrow we will continue the personalized theme and cover another mind blowing talk that demonstrated how far we really have to go before we can possibly expect to see major shifts in outcome based on the underlying biology and matching appropriate targeted therapy.








 Jeffrey Engelman from MGH persuasively presented on why we need combination therapies to overcome resistance. He noted that:
Jeffrey Engelman from MGH persuasively presented on why we need combination therapies to overcome resistance. He noted that: One enduring image I have from the plenary presentation on “The Genetic Basis for Cancer Therapy” by Bill Sellers, VP/Global Head Oncology at Novartis Institutes for BioMedical Research was the video he showed of the robots that are used for automated cell profiling.
One enduring image I have from the plenary presentation on “The Genetic Basis for Cancer Therapy” by Bill Sellers, VP/Global Head Oncology at Novartis Institutes for BioMedical Research was the video he showed of the robots that are used for automated cell profiling. To do this, Sellers described how Novartis have built a robotic system that e.g. automates cell profiling. In approx 3 months with this system we can profile 600 cell lines for about 1500 compounds, he said.
To do this, Sellers described how Novartis have built a robotic system that e.g. automates cell profiling. In approx 3 months with this system we can profile 600 cell lines for about 1500 compounds, he said.